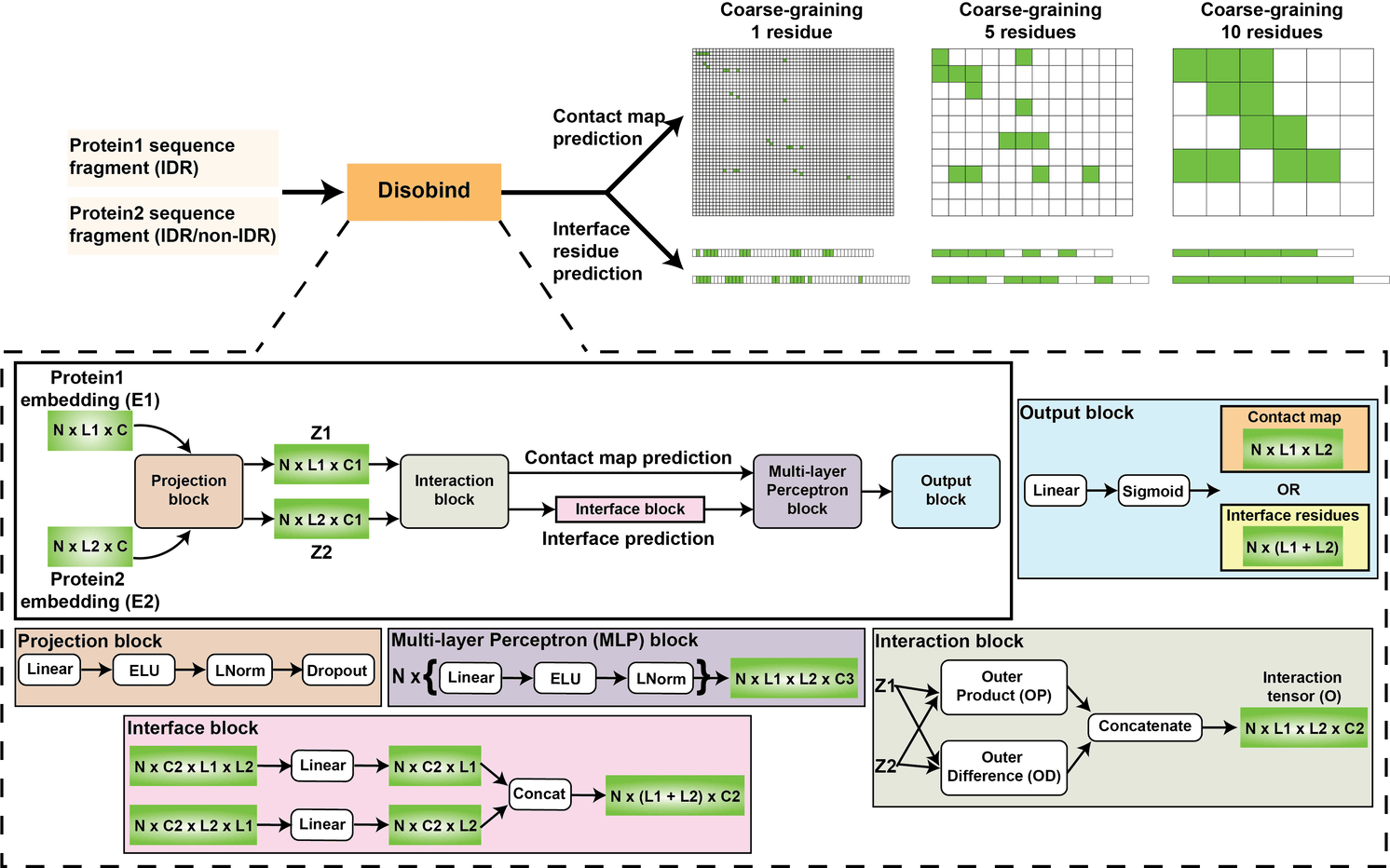
A deep learning method for predicting interactions for intrinsically disordered regions of proteins
K. Majila, V. Ullanat, S. Viswanath
Cell Systems,
2026
Recent Methods from Statistical Inference for Integrative Structural Modeling
S. Arvindekar*, K. Majila*, S. Viswanath
Machine Learning, Artificial Intelligence, and Big Data, Springer Handbook of Chem- and Bioinformatics,
2026
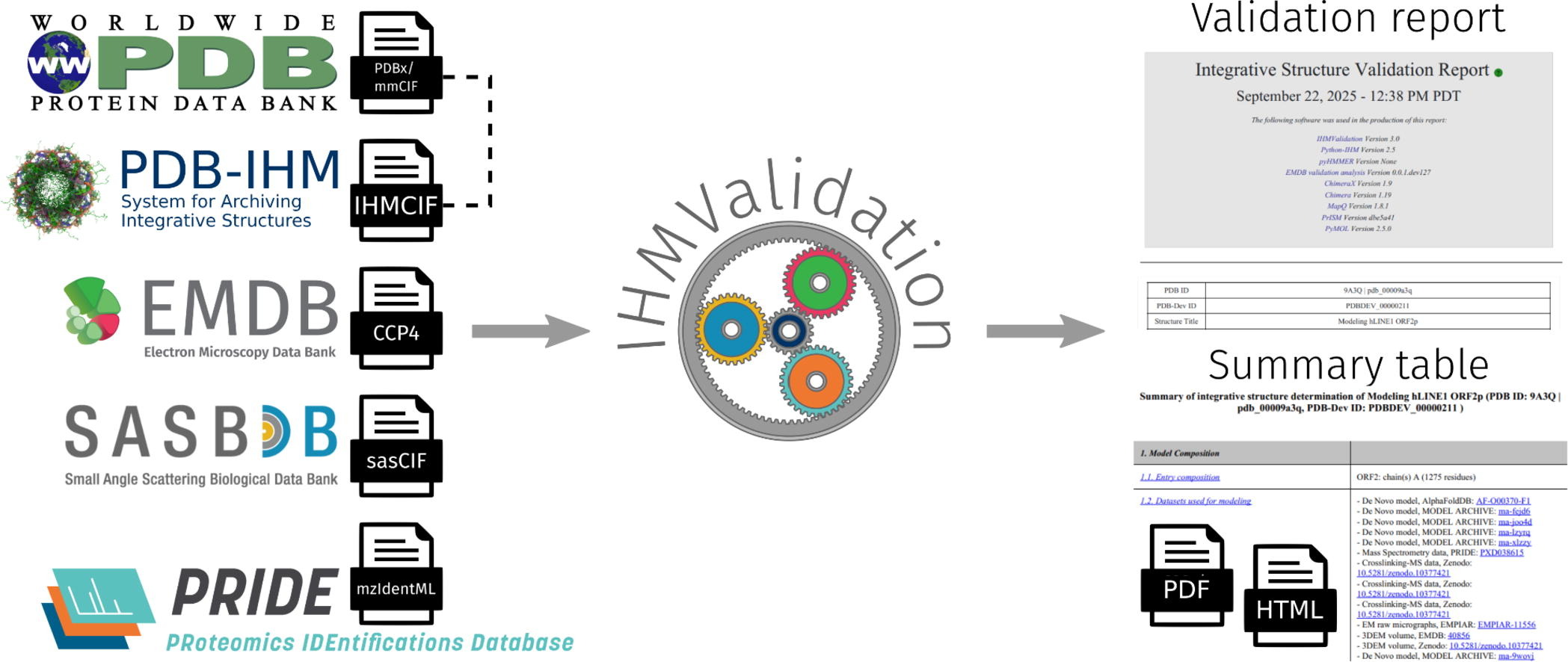
IHMValidation: Assessment of Integrative Structure Models Deposited to the Protein Data Bank
A. Zalevsky, B. Vallat, B. M. Webb, H. Tangmunarunkit, M. Sekharan, A. Shafaeibejestan, S. Ganesan, J. Sagendorf, C. M. Jeffries, J. Trewhella, A. Graziadei, J. A. Vizcaíno, A. Leitner, J. Rappsilber, E. Peisach, J. W. Flatt, J. Y. Young, K. Majila, S. Viswanath, C. Kesselman, J. C. Hoch, G. Kurisu, K. Morris, S. Velankar, H. M. Berman, S. K. Burley, A. Sali
Journal of Molecular Biology,
2025
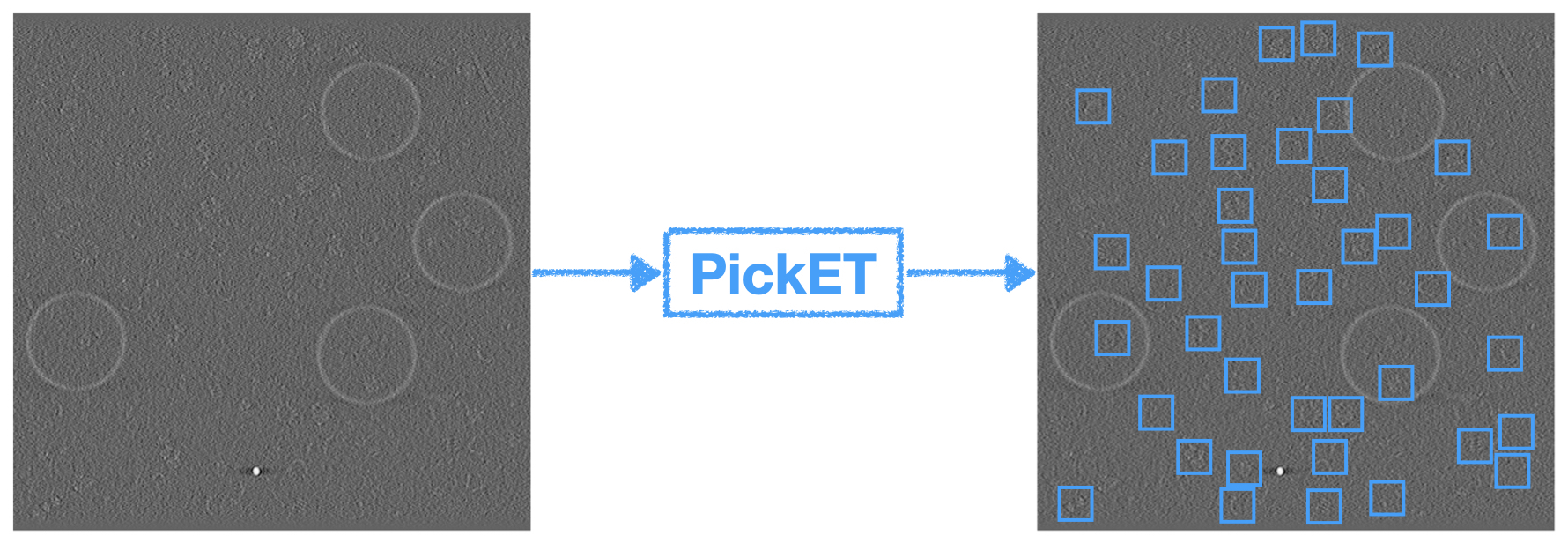
PickET: An unsupervised method for localizing macromolecules in cryo-electron tomograms
S. Arvindekar, O. Golatkar, S. Viswanath
bioRxiv,
2025
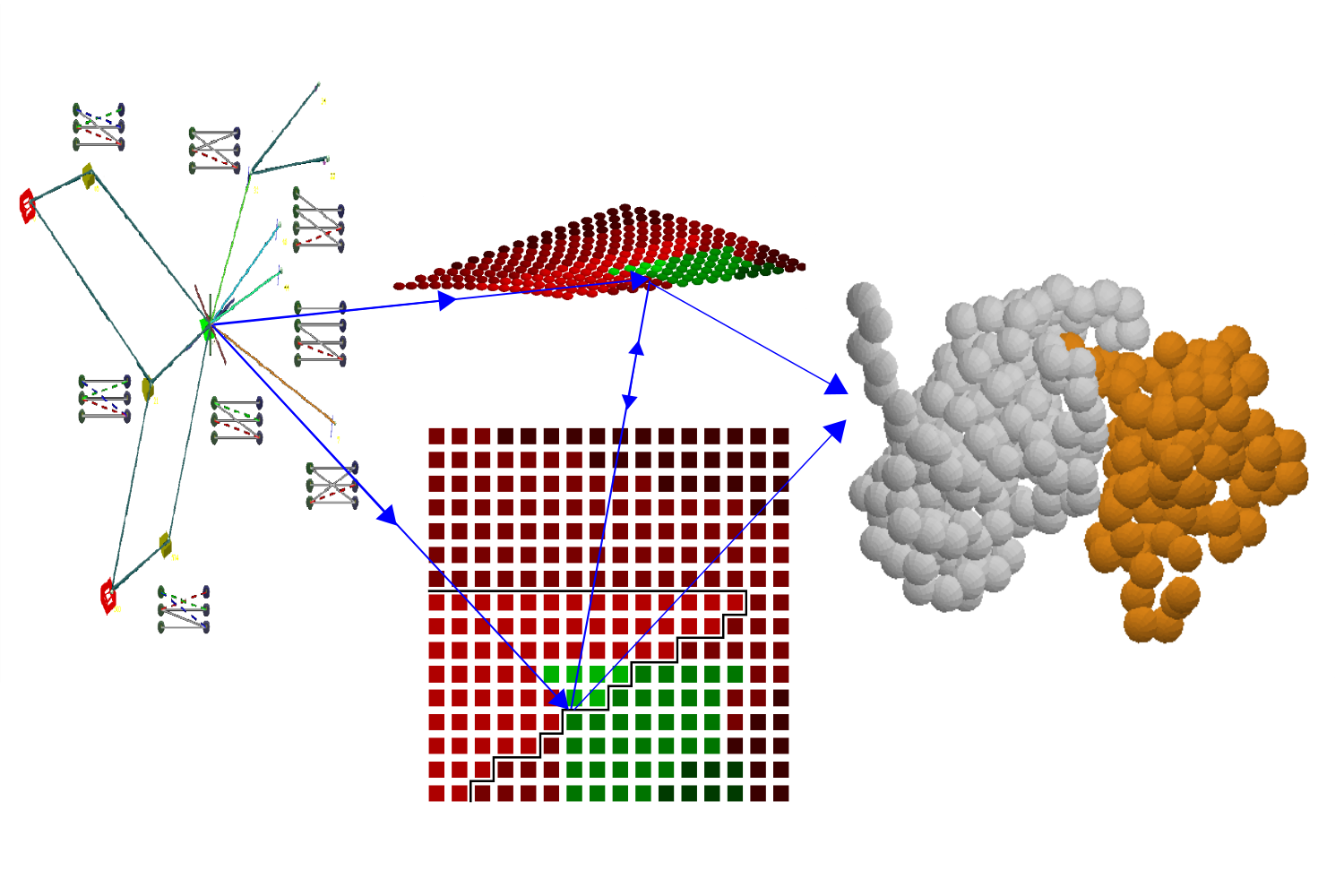
A new discrete-geometry approach for integrative docking of proteins using chemical crosslinks
Y. Zhang*, M. Jindal*, S. Viswanath#, M. Sitharam#
Journal of Chemical Information and Modeling,
2025
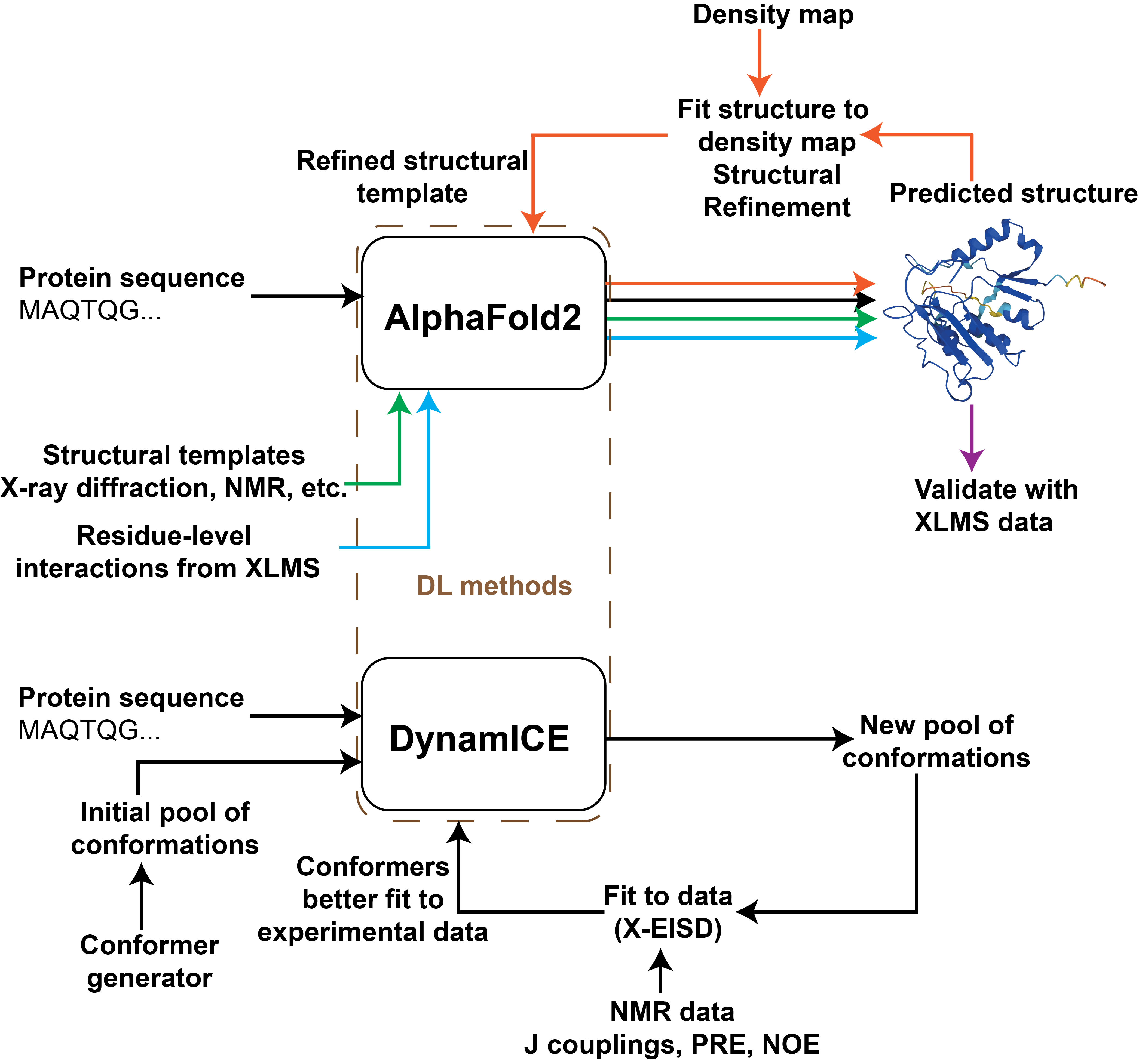
Frontiers in integrative structural biology: modeling disordered proteins and utilizing in situ data
K. Majila*, S. Arvindekar*, M. Jindal, S. Viswanath
Quarterly Reviews of Biophysics Discovery (invited Perspective),
2025
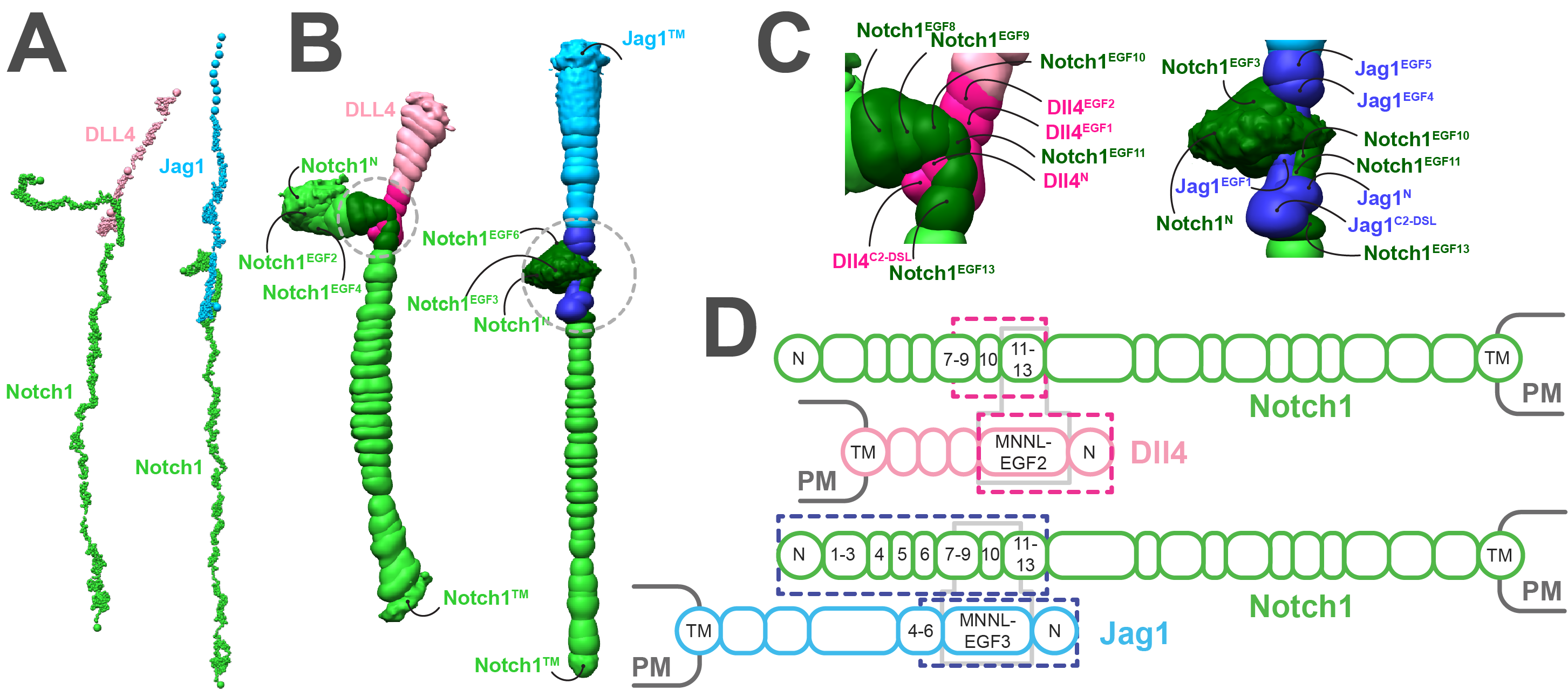
Multiscale simulations reveal architecture of NOTCH protein and ligand specific features
S. Rathore, D. Gahlot, J. Castin, A. Pandey, S. Arvindekar, S. Viswanath, L. Thukral
Biophysical Journal,
2025
Alphafold opens the doors to deorphanizing secreted proteins
S. Viswanath
Cell Systems (invited Preview),
2024
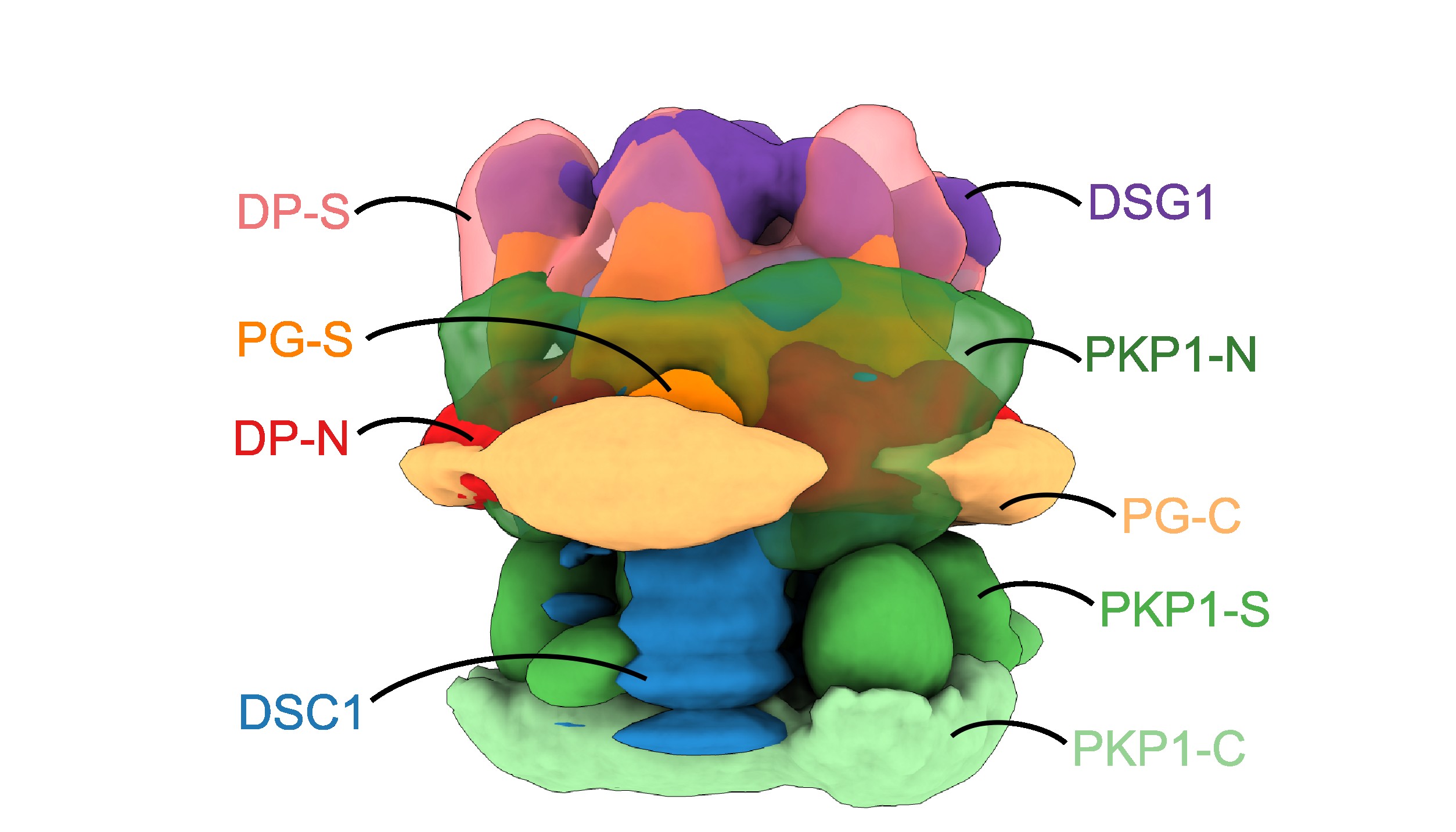
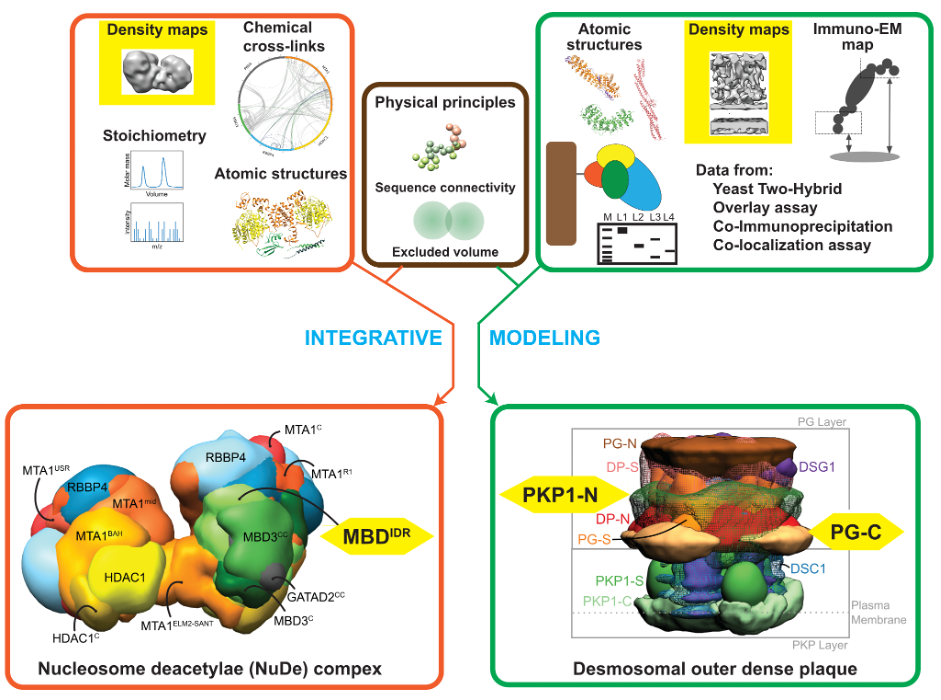
The molecular architecture of the desmosomal outer dense plaque by integrative structural modeling
S. Pasani, K. Menon, S. Viswanath
Protein Science,
2024
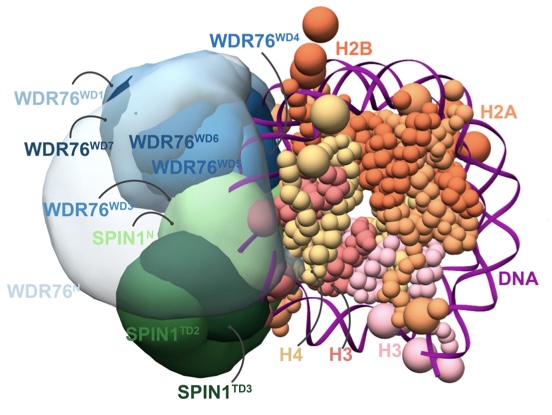
An integrated structural model of the DNA damage-responsive H3K4me3 binding WDR76:SPIN1 complex with the nucleosome
X. Liu, Y. Zhang, Z. Wen, Y. Hao, C. A.S. Banks, J. Cesare, S. Bhattacharya, S. Arvindekar, J. J. Lange, Yixuan Xie, B. A. Garcia, B. D. Slaughter, J. R. Unruh, S. Viswanath, L. Florens, J. L. Workman, and M. P. Washburn
PNAS,
2024
Optimizing representations for integrative structural modeling using Bayesian model selection
S. Arvindekar, A. S. Pathak, K. Majila, S. Viswanath
Bioinformatics,
2024
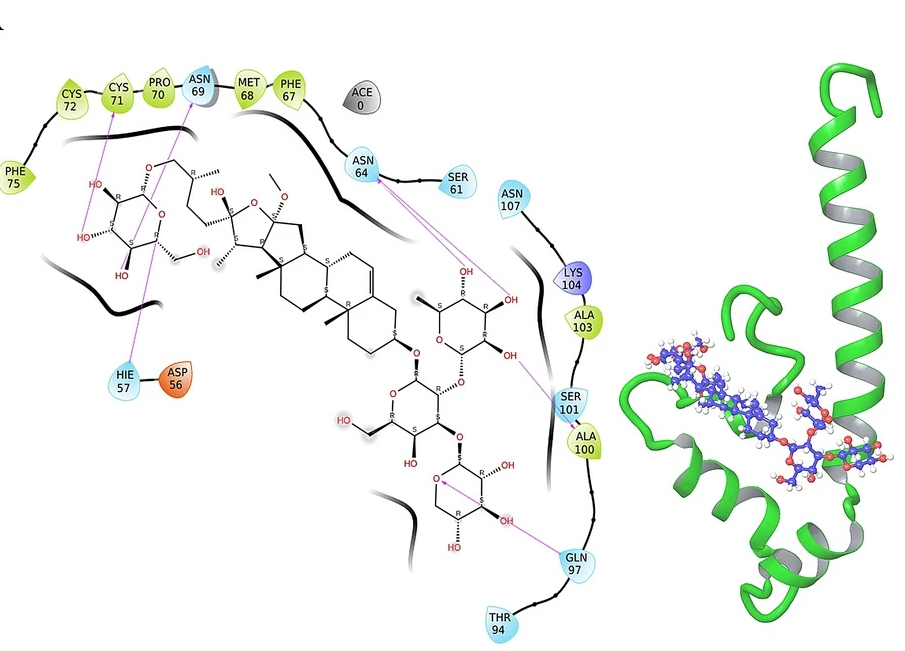
Identification of potential modulators of IFITM3 by in-silico modeling and virtual screening
V. Tiwari and S. Viswanath
Scientific Reports,
2022
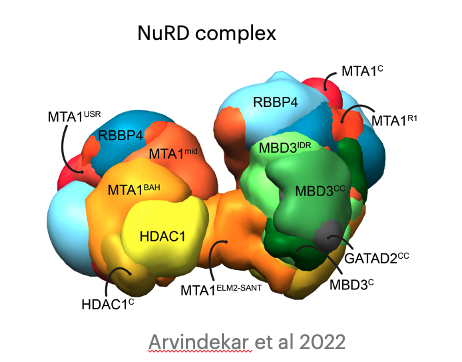
Molecular architecture of nucleosome remodeling and deacetylase sub-complexes by integrative structure determination
S. Arvindekar, M. J. Jackman, J.K.K. Low, M.J. Landsberg, J.P. Mackay, S. Viswanath
Protein Science,
2022
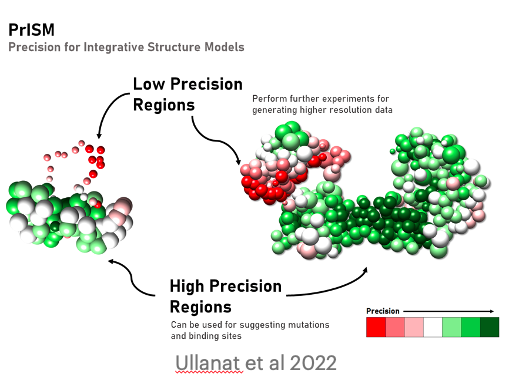
PrISM: precision for integrative structural models
V. Ullanat, N. Kasukurthi, S. Viswanath
Bioinformatics,
2022
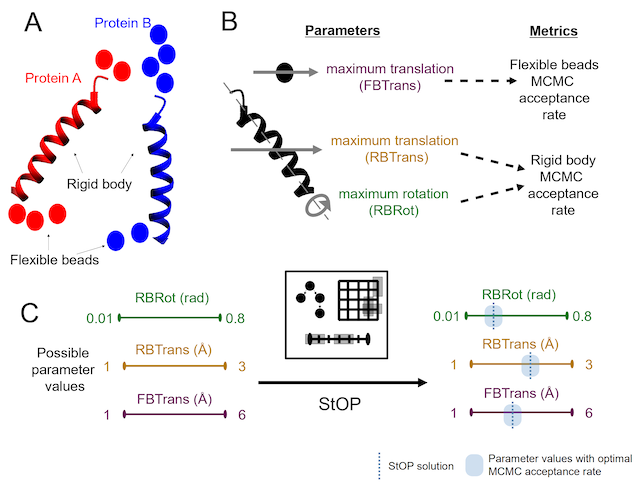
A Framework for Stochastic Optimization of Parameters for Integrative Modeling of Macromolecular Assemblies
S. Pasani, S. Viswanath
Life (Spl. Issue on Computational Modeling of Kinetics in Biological Systems),
2021
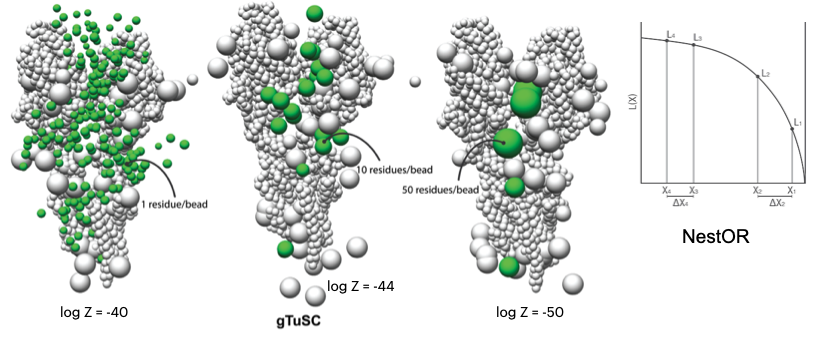
CM1-driven assembly and activation of Yeast γ-Tubulin Small Complex underlies microtubule nucleation
A. Brilot, A.S. Lyon, A. Zelter, S. Viswanath, A. Maxwell, R. Johnson, K.C. Yabut, M. MacCoss, E. Muller, A. Sali, T.N. Davis, D. A. Agard
eLife,
2021
Using Integrative Modeling Platform to compute, validate, and archive a model of a protein complex structure
D. Saltzberg, S. Viswanath, I. Echeverria, I.E. Chemmama, B. Webb, A. Sali
Protein Science,
2021
Modeling biological complexes using Integrative Modeling Platform, Biomolecular Simulations Series
D. Saltzberg, C.H. Greenberg, S. Viswanath, I.E. Chemmama, B. Webb, R. Pellarin, I. Echeverria, A. Sali
Methods in Molecular Biology,
2019
Optimizing model representation for integrative structure determination of macromolecular assemblies
S. Viswanath and A. Sali
PNAS,
2019
Integrative structure modeling with the Integrative Modeling Platform
B. Webb, S. Viswanath, M. Bonomi, R. Pellarin, C.H. Greenberg, D. Saltzberg, A. Sali
Protein Science,
2018
Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures
S. Viswanath, I.E. Chemmama, P. Cimermancic, A. Sali
Biophysical Journal,
2017
The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling.
S. Viswanath, M. Bonomi, S.J. Kim, V.A. Klenchin, K.C. Taylor, K.C. Yabut, N.T. Umbreit, H.A. Van Epps, J. Meehl, M.H. Jones, D. Russel, J.A. Velazquez-Muriel, M. Winey, I. Rayment, T.N. Davis, A. Sali, E.G. Muller
Molecular Biology of the Cell,
2017
Prediction of homoprotein and heteroprotein complexes by protein docking and template-based modeling: A CASP-CAPRI experiment
M.F. Lensink, S. Velankar, …, S. Viswanath, R. Elber, …, (several authors), S.J. Wodak
Proteins,
2016
Extension of a protein docking algorithm to membranes and applications to amyloid precursor protein dimerization
S. Viswanath, L. Dominguez, L.S. Foster, J.E. Straub, R. Elber
Proteins,
2015
Architecture of the human and yeast general transcription and DNA repair factor TFIIH
J. Luo, P. Cimermancic, S. Viswanath, C. C. Ebmeier, B. Kim, M. Dehecq, V. Raman, C.H. Greenberg, R. Pellarin, A. Sali, D. Taatjes, S. Hahn, J. Ranish
Molecular Cell,
2015
DOCK/PIERR: web server for structure prediction of protein-protein complexes
S. Viswanath, D.V.S. Ravikant, R. Elber
Protein Structure Prediction, Methods in Molecular Biology,
2014
Improving ranking of models for protein complexes with side chain modeling and atomic potentials
S. Viswanath, D.V.S. Ravikant, R. Elber
Proteins,
2013
Analyzing milestoning networks for molecular kinetics: Definitions, algorithms, and examples
S. Viswanath, S.M. Kreuzer, A.E. Cardenas, R. Elber
The Journal of Chemical Physics,
2013