ISB Lab Software and Resources
Check out software developed by our group and other open-source freebies at ISB Lab GitHub.
IDRs in complexes
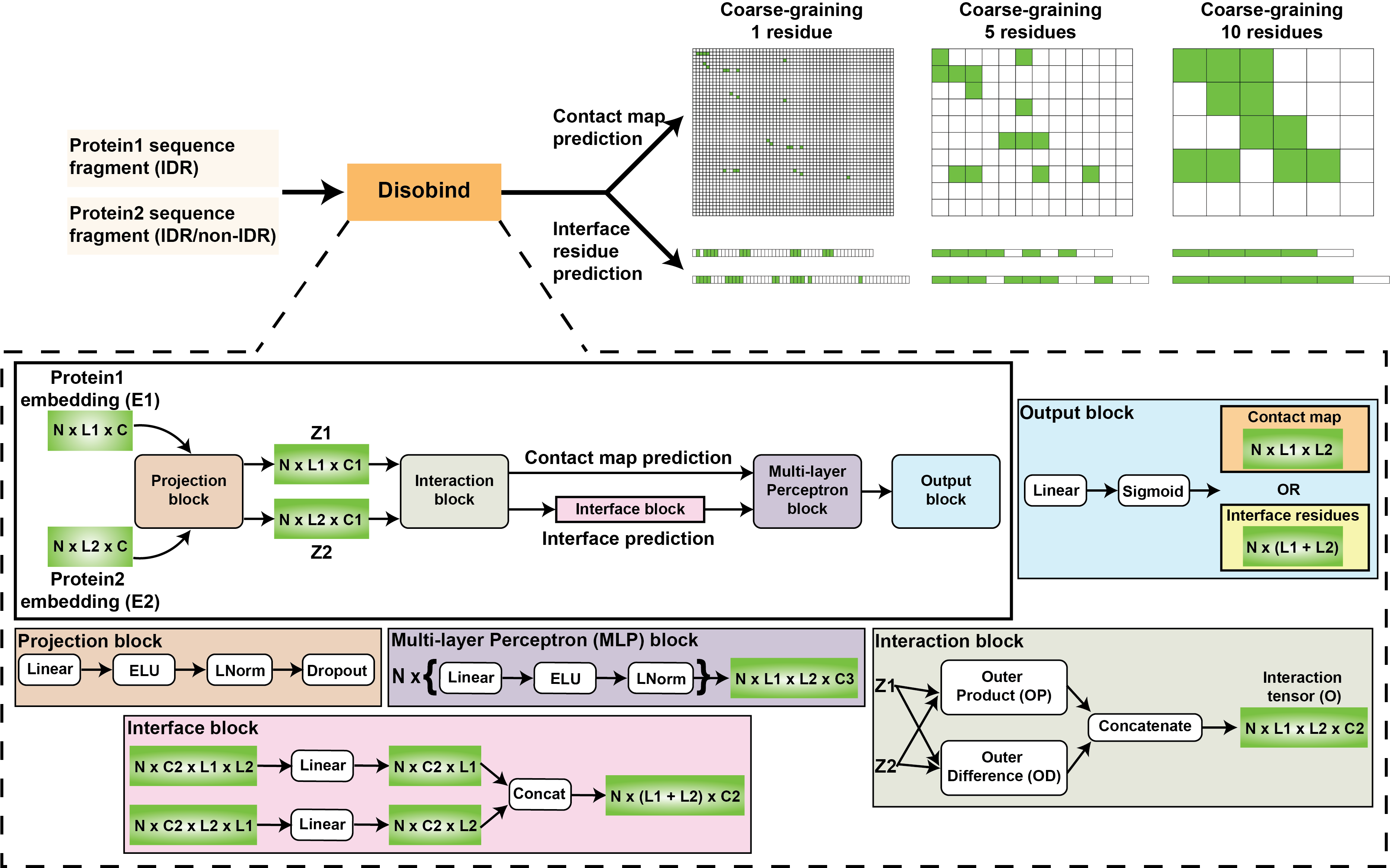
Disobind
A deep-learning method to predict inter-protein contact maps and interface residues for an intrinsically disordered region (IDR) and a partner protein from their sequences
Cryo-electron tomography

Integrative structural modeling
Wall-EASAL
A benchmark of hetero-dimeric complexes, with component atomic structures and chemical crosslinks used to compare integrative modeling methods: Wall-EASAL and IMP.
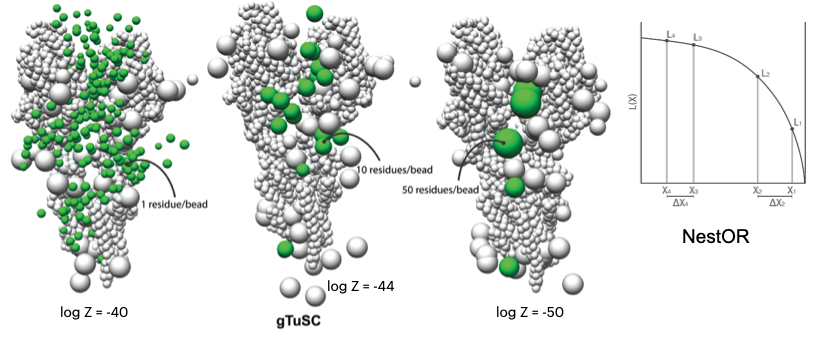
NestOR
A Python module to perform Nested sampling-based optimization of representation for integrative structural modeling.
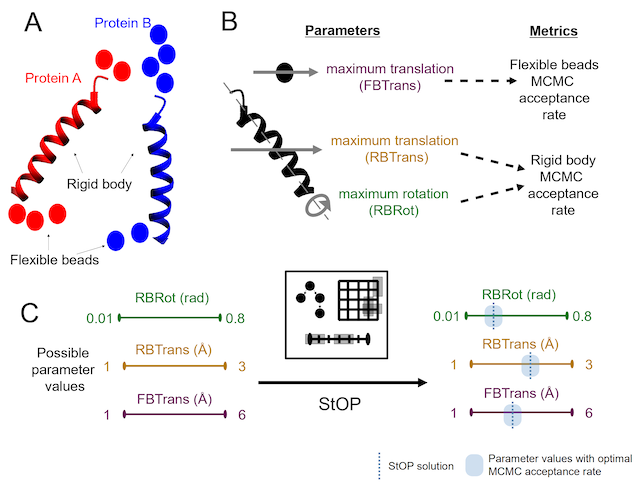
StOP
A gradient-free, parallel, global, stochastic, multi-objective optimization algorithm primarily written to optimize the Markov Chain Monte Carlo (MCMC) sampling parameters for the Integrative Modeling Platform (IMP).
Sampcon
Pipeline for analyzing integrative models after Markov Chain Monte Carlo (MCMC) sampling. Includes tests for assessing sampling exhaustiveness, clustering models, and calculating precision.

Courses and Workshops
Integrative Modeling Tutorials and Workshops
- IMP Tutorial at EMBOCEM3DIP IISc Bangalore 2024. GitHub | Talks
- IMP Tutorial at MASFE IISER Pune 2023. GitHub | Talks
- IMP Workshops
Graduate Courses at NCBS
- Statistical Inference in Biology (internal link) co-taught with Dr Shaon Chakrabarti at NCBS.
- Crash course in Python taught during the Statistical Inference course.
