StrIDR
Structures of Intrinsically Disordered Regions
Version: 1.0 Release_date: 08_2024
A database of intrinsically disordered regions of proteins (IDRs) resolved in structure.
StrIDR is a database of IDRs, confirmed via experimental or homology-based evidence, that are resolved in experimentally determined structures.
Existing databases provide extensive information for IDRs at the sequence level. However, only a tiny fraction of these IDRs are associated with an experimentally determined protein structure. Moreover, the disordered region of interest could be unresolved even if a structure exists.
StrIDR is expected to be useful for gaining insights into the dynamics, folding, and interactions of IDRs. It may provide structural data for computational methods to studying IDRs including molecular dynamics (MD) simulations and machine learning (ML).
We obtain disorder annotations, sequence, and structure information from the following databases:









Statistics
Experimental techniques for structure determination of IDR proteins in StrIDR.
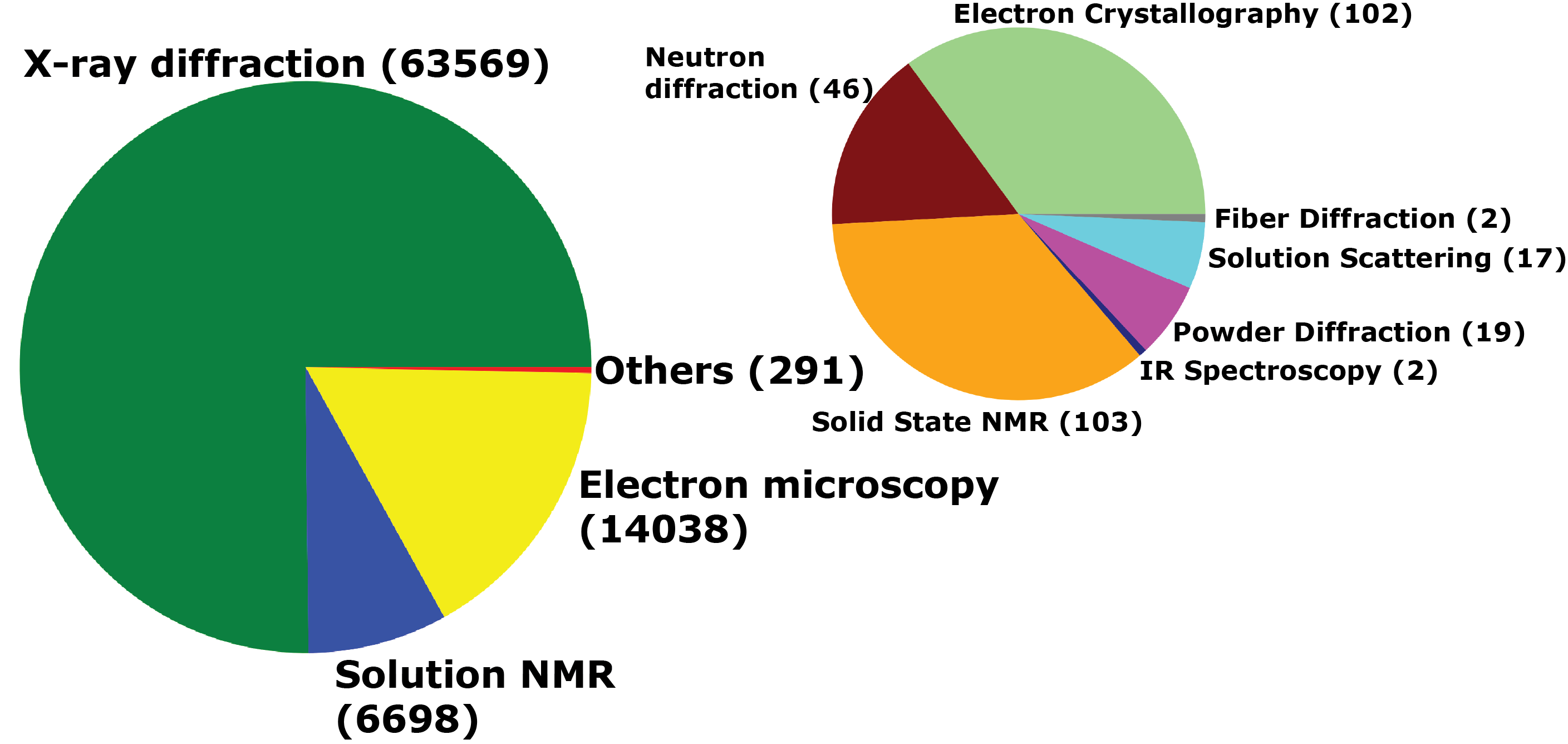
StrIDR contains 84598 PDB entries 303397 IDR chains 22508 Unique UniProt accessions.
How to Cite
Majila K., Viswanath S. (2024) StrIDR: a database of intrinsically disordered regions of proteins with experimentally resolved structures.
bioRxiv doi: https://doi.org/10.1101/2024.08.22.609111
Contact us at: shruthiv AT ncbs DOT res DOT in
Data access
 This work is licensed under a
Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
This work is licensed under a
Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
