ISB Lab Software and Resources
Check out software developed by our group and other open-source freebies at ISB Lab GitHub.
| Name | Description | Link | |
|---|---|---|---|
| Disobind | 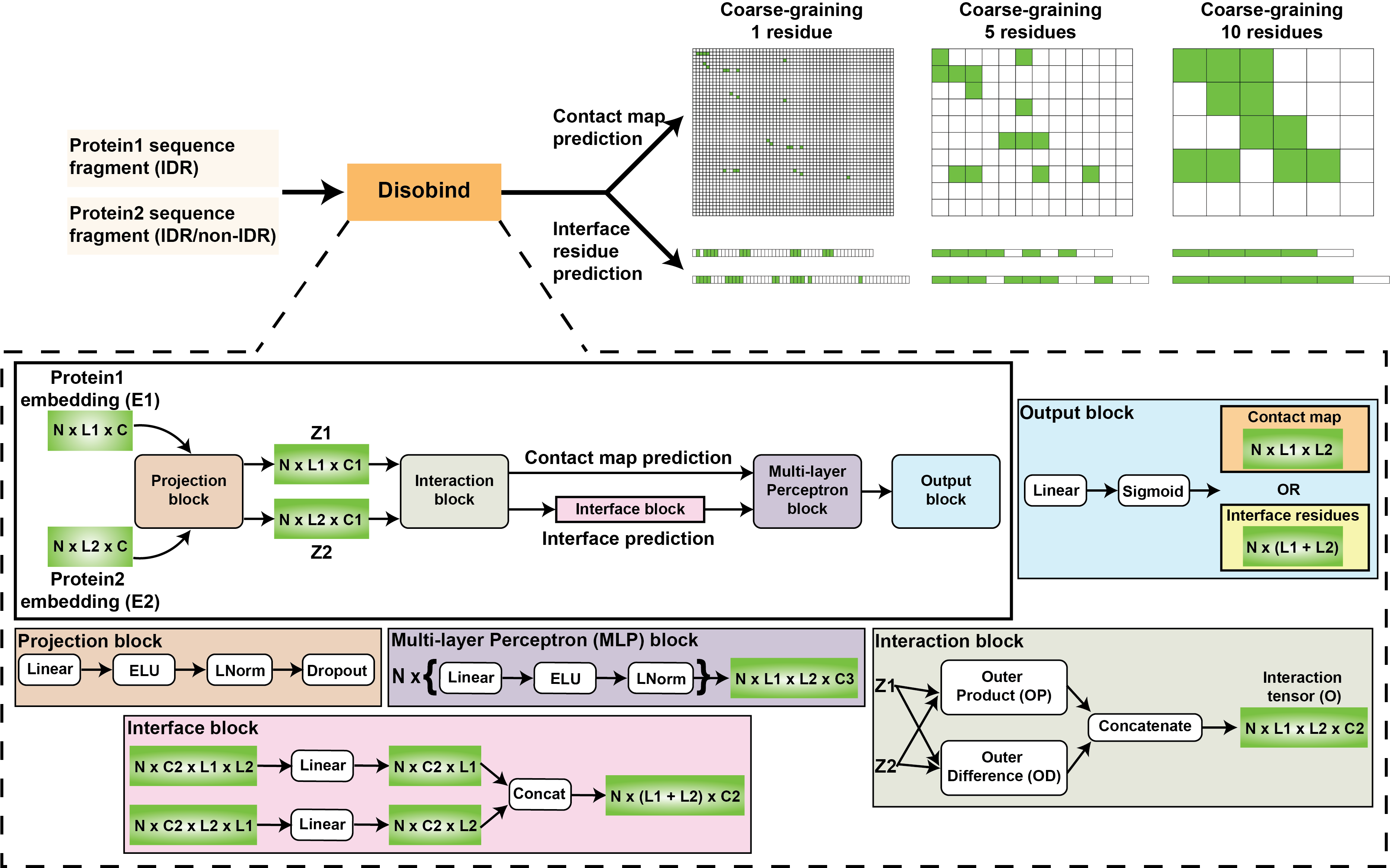 |
A deep-learning method to predict inter-protein contact maps and interface residues for an IDR and a partner protein from their sequences |
GitHub |
| Wall-EASAL |  |
Integrative docking with crosslinks. |
GitHub EASAL Software |
| NestOR | 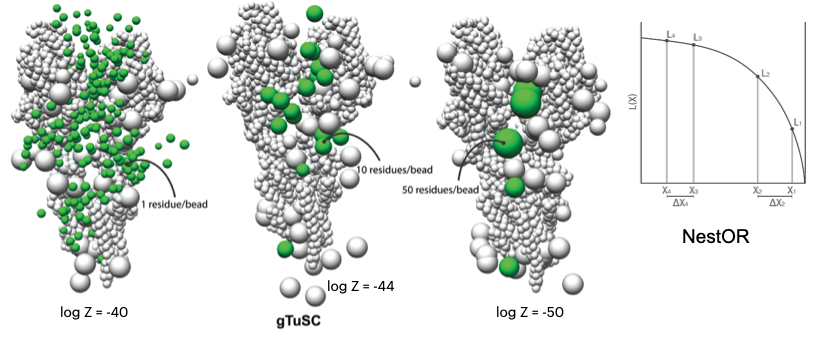 |
Nested sampling-based optimization of representation for integrative structural modeling. | GitHub |
| PrISM |  |
Package to annotate high- and low-precision regions in integrative structure models. Now used by wwPDB to validate integrative models! | GitHub |
| StOP | 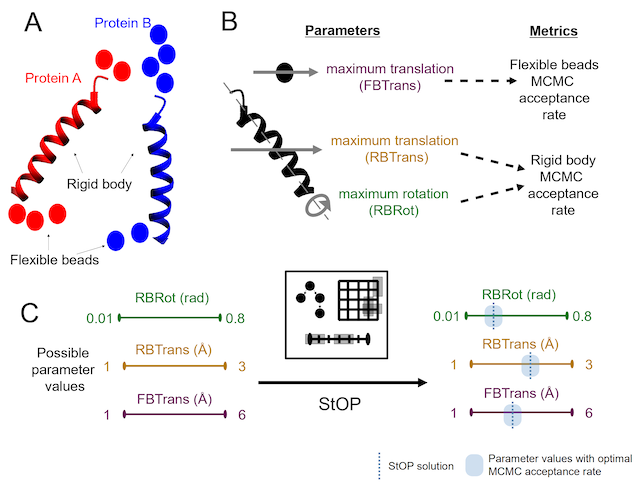 |
Stochastic Optimization of Parameters (StOP) automates the tuning of MCMC parameters for stochastic sampling in IMP. - Check this tutorial for optimizing Monte Carlo move sizes/restraint weights. - And this tutorial for optimizing replica exchange temperatures. |
GitHub |
| Sampcon |  |
Pipeline for analyzing integrative models after MCMC sampling. Includes tests for assessing sampling exhaustiveness, clustering models, and calculating precision. | GitHub |
| Scoring functions for protein-protein docking and other utilities for parsing PDBs |  |
PISA (atomic), PIE (residue), and C3 (combination) scoring functions for ranking models. Interface RMSD calculation, utilities for modifying PDBs, adding chain names, and running Modeller. | GitHub |
Courses and Workshops
Integrative Modeling Tutorials and Workshops
- IMP Tutorial at EMBOCEM3DIP IISc Bangalore 2024. GitHub | Talks
- IMP Tutorial at MASFE IISER Pune 2023. GitHub | Talks
- IMP Workshops
Graduate Courses at NCBS
- Statistical Inference in Biology (internal link) co-taught with Dr Shaon Chakrabarti at NCBS.
- Crash course in Python taught during the Statistical Inference course.